

A Research Project by Jakob H. Waterborg, Ph.D.
© 2007-2010. All rights reserved.
Permission is required for use of any unpublished data.
The supplementary files of this publication contain all curated histone sequences, alignments and deduced phylogenies.
-
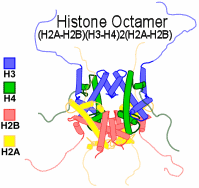
1. Histone H3 is present twice in the center of the nucleosome, the protein octamer that packages DNA.[1]
-
2. Histone H3 proteins (135 amino acids) exist characteristically as 2 variant forms
-
with very few sequence differences,
and they function differently in nucleosome assembly.
Function-related sequences differences exist in residues 31, 41, 87, 89 and 90 (see examples below). [2-4]
-
3. Replacement RI histone H3
- Synthesis is not restricted to S phase, i.e. it is named "RI" for Replication-Independent. [3]
Synthesis is often constitutive or is induced upon tissue differentiation. [2] - Assembly into nucleosomes is observed across transcribed genes, denuded of histones. [3-4]
This leads to the characteristic replacement of existing histone H3 protein by RI H3 forms. [2] - The transcription-coupled nucleosome assembly machinery selectively uses only RI histone H3. [3]
- In animals (metazoa), Replacement H3 variants are known as H3.3
- In plants, Replacement H3 variants are typically named H3.2
-
4. Replication-Coupled histone H3
- Synthesis is restricted to S phase, i.e. it is "RC" for Replication-Coupled. [3]
Typically, RC H3 gene transcription is repressed upon completion or inhibition of DNA synthesis. [5] - Assembly into new nucleosomes across replicated DNA sequences.
The RC assembly machinery will incorporate RC and RI histone H3 variants alike. [3]- In animals (metazoa), RC histone H3 variants are known as H3.1/H3.2
- In plants, RC histone H3 variants are often named H3.1
|
 (Variant-function characteristic differences are capitalized; pairwise differences are bolded; stars mark 6 acetylatable lysines which reduce acid-urea gel mobility.)
(Variant-function characteristic differences are capitalized; pairwise differences are bolded; stars mark 6 acetylatable lysines which reduce acid-urea gel mobility.)

-
5. Single chimeric histone H3 protein in Ascomycetes
- In ascomycetes like Saccharomyces cerevisiae only a single H3 form exists. [6]
- The ascomycete H3 protein looks like a chimera with #87 as in metazoan and plant RC forms
and with #31 and #89-90 as in higher RI variants. - In Schizosaccharomyces pombe a similar chimeric H3 protein exists:
- It is made in S phase from 2 cell cycle regulated loci, and
- it is produced constitutively from a third locus. [7]
- Distinct RC and RI nucleosome assembly pathways still exist in these ascomycetes. [8-10]
- This suggests that in ascomycetes the multiplicity of RC and RI H3 proteins was reduced by retention of a dual functionality in a single protein.

-
6. Two histone H3 variants in Basidiomycetes
- In basidiomycetes like the corn smut Ustilago maydis two distinct H3 genes exist, each as a single-copy gene within the fully sequenced genome.
- We have purified and separated the two histone H3 proteins from whole cells by guanidine.HCl sonication extraction, Biorex-70 recovery, reversed-phase hplc (Fig. 1) and acid-urea-Triton (AUT) gel electrophoresis (Fig. 2).
- [35S]-Methionine labeling in vivo allowed identification of the two protein forms as products of genes U1 and U2: only one of the two H3 species is labeled by methionine, as predicted from the protein sequence (Table 3). This identifies the higher-mobility H3.2 form as the protein product of the U2 locus (Fig. 3).
- The sequence differences between the U1 and U2 forms (Table 3), when compared with animal (Table 1) and plant (Table 2) histone H3 forms, including extensive phylogenetic explorations, suggest that U1/H3.1 with a characteristic H3 motif of 87-89 90 = S-I G is the RI form and that U2/H3.2 with the S-V M motif is the replication-coupled RC form.
|
-
7. Functional RI and RC identification of Ustilago H3 variants
- Assembly of the Replacement H3 across nucleosome-depleted regions of actively transcribed genes predicts, as demonstrated for animal H3.3 and plant H3.2 RI replacement histones [12], predicts that H3.1 will have a higher level of acetylation with faster turnover rates. This has been demonstrated for Ustilago H3.1 (Fig. 3-4)
- If expression of gene U2 into H3.2 protein produces a Replication-Coupled H3 protein, one would expect a high level of H3.2 synthesis in S phase and a low level of synthesis outside of S-phase. Conversely, expression of the U1 gene into H3.1 protein is expected to be less clearly cell cycle regulated. Preliminary observations in synchronized cells (Fig. 5) and an increase in the relative abundance of the putative RI-H3.1 variant in non-proliferating cultures (Fig. 6) supports the H3.1=RI and H3.2=RC identification.
- Transcriptional replacement of pre-existing histone (i.e. S-phase deposited RC histone H3.2) across transcribed genes by the suspected RI H3.1 form of H3 predicts that the protein stability of H3.1 is less than that of histone H3.2.
-
The differential H3 variant turnover, measured by pulse-chase experiments, provided major proof that alfalfa histone H3.2 was a Replacement histone. [2]
- This prediction is experimentally being tested by measuring the specific radioactivity decay for both H3 forms during a chase after pulse-labeling of an asynchronous log culture of Ustilago cells with [3H]-lysine.
This experiment may not yield definitive results because only limited amounts of tritiated label could be incorporated in the pulse period of 30 min.
In view of this experimental limitation, the following experiments are planned.-
They will measure the differential incorporation of U1/H3.1 and U2/H3.2 smut H3 proteins into distinct chromatin environments, and they will measure the subsequent stability of the incorporated H3 variant proteins.
- This prediction is experimentally being tested by measuring the specific radioactivity decay for both H3 forms during a chase after pulse-labeling of an asynchronous log culture of Ustilago cells with [3H]-lysine.
- Measure the absolute rate of H3 variant specific protein turnover by gene constructs in which U1 and U2 coding sequences have been fused to Green and/or Red Fluorescent Protein sequences under control of the cadmium-inducible metallothionein promoter. Removal of the heavy metal induction by exchange of the growth medium will start the chase condition because the MTT promoter will return to its default repressed state [12].
Transformation of Ustilago will be used to stably integrate these constructs in the smut genome without affecting the endogenous single U1 and U2 loci. Single and combined transformation cell lines will be established to measure directly and relative to eachother the synthesis and turnover of each gene product during log and stationary phase growth, and in cell cycle-synchronized cultures. - As controls, smut transformations will be performed with the U1 and U2 coding sequences - with or without fluorescent tag - under control of their natural promoter or under control of the promoter of the other variant. This will show whether the predicted S-phase expression of U1 and predicted constitutive expression of U2 rather than the predicted differences in histone proteins are determinants for RC vs RI functionality. In Tetrahymena, to some degree both factors appear to play a role [12].
-
With their natural promoters, normal growth is expected with the opportunity to assess the effects of the fluorescent protein extension in Ustilago. It is known that C-terminal GFP extension of histone H3 is without significant effect [12].
Upon promoter exchange, the effects of inappropriate expression of distinct H3 forms may be limited to reduced growth rates caused by limiting availability of histone H3 for chromatin replication or to maintain chromatin packaging of transcribed genes, or may be severe and lethal. - In a parallel set of experiments, GFP-tagged or immuno-tagged (e.g. by poly-His) U1 or U2 gene constructs with their natural or with the exchanged promoter, will be targeted to the endogenous U1 and/or U2 loci for gene replacement of the endogenous copy. As with the GFP-H3 add-on experiments, cell viability, growth rate, cell cycle progression and relative incorporation into bulk, newly replicated or transcribed chromatin domains will be measured.
-
This experiment will determine in a more rigorous way than the one described above, the essential contribution that RI and RC histone H3 variants make in the nucleosome assembly of newly replicated or across transcribed chromatin.
Analysis methods will include assessment of H3 dynamic and steady-state acetylation levels in AUT gel electrophoresis by stainging and fluorography, confocal microscopic observation of cells, western blotting of histone H3 preparations from AUT and SDS gels (in which GFP-H3 forms will separate due to their increased size from endogenous H3 gene products), and various forms of ChIP methodologies in which acetylated-lysine site-specific antibodies and H3-immuno tag antibodies will be used as employed by others [xxx].
-
Specific Aim 1: demonstrate that U1 product H3.1 = RI and that U2 product H3.2 = RC by evaluating the following testable predictions.
-
8. Specific Aim 2: Identification of histone H3 residues that define RI and RC function
- Modify existing histone H3 yeast plasmids by site-directed mutagenesis to create, in-part or completely, the smut H3.1 and H3.2 genes, tailed by a defined immuno sequence tag, and determine bulk and sequence-specific incorporation by standard ChIP (Chromatin Immuno Precipitation) analysis of yeast.
-
Residues targeted for change are first of all amino acids 31, 89 and 90, which differ between yeast and smut H3 forms and which have been implicated in RC/RI functional differences. [3]
Residue 87 will be modified as in animal and plant H3 variants to define its contribution.
The effect of changing tyrosine 41 to phenylalanine, a difference between H3 variants in plants that could contribute to RC-RI functional differences, will be tested but is likely to show no effect, as predicted by phylogenetic analysis.
Residues 95 and 135 will be included even though there is no indication from phylogenetic comparisons that functional H3 variant differences may reside here. Variability at or near residue 95 is relatively high - as far as H3 variability goes - as is the variability of the terminal H3 residue across broad phylogenetic branches. This suggests that functional selective pressure on these two residues is probably lower than anywhere else in histone H3 (see the phylogenetic analysis on this website) and is unlinked to RC-RI functional differences. - Determine the incorporation of the smut H3 variants relative to the yeast H3 histone by ChIP analysis across well studied DNA domains in yeast under selected conditions such as log growth, arrested or stationary cultures.
-
Expected outcomes:
- RC H3 will only incorporate in newly replicated DNA, assembled by the replication nucleosome assembly pathway. [3,8]
- RI H3 will be incorporated in growing and arrested cultures alike, with a clear preference for transcribed sequences where it will be incorporated into replacement nucleosomes by the transcription-linked chromatin repair machinery. [2-3,8]
- Histone H3 constructs with some but not all RC- or RI-specific residues may show intermediate behaviors in this assay or may be excluded from chromatin assembly.
- Test the essential characteristics of any smut-H3-like histone form directly as rescue plasmids - singly or in combinations of sequence variations - by transformation of yeast strains in which both histone H3 gene copies have been deleted. [13]
-
Expected outcome:
- Transformed yeast cultures will only survive and grow if the plasmid-encoded H3 protein - or combination of proteins - provides all the essential functions that reside in the putative chimeric yeast H3 histone.
-
For this experimental exploration, the yeast Saccharomyces cerevisiae will be used, transforming it by Ustilago maydis (smut) H3 genes and their sequence derivatives.
References
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. "Crystal structure of the nucleosome core particle at 2.8Å resolution." Nature 389, 251-260, 1997.
- Waterborg JH. "Histone synthesis and turnover in alfalfa. Fast loss of highly acetylated replacement histone H3.2" J. Biol. Chem. 268, 4912-4917, 1993.
- Ahmad K, Henikoff S. "Histone H3 variants secify modes of chromatin assembly." PNAS 99, 16477-16484, 2002.
- Waterborg JH, Robertson AJ. "Common features of analogous replacement histone H3 genes in animals and plants." J. Mol. Evol. 43, 194-206, 1996.
- Kapros T, Robertson AJ, Waterborg JH. "Histone H3 transcript stability in alfalfa." Plant Mol. Biol. 28, 901-914, 1995.
- Smith MM, Andresson OS. "DNA sequences of yeast H3 and H4 histone genes from two non-allelic gene sets encode identical H3 and H4 proteins." J. Mol. Biol. 3, 663-690, 1983.
- Yu LL, Gorovsky MA. "Constitutive expression, not a particular primary sequence, is the important feature of the H3 replacement variant hv2 in Tetrahymena thermophila." Mol. Cell. Biol. 17, 6303-6310, 1997.
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. "Dynamics of replication-independent histone turnover in budding yeast." Science 315, 1405-1408, 2007.
- Choi ES, Shin JA, Kim HS, Jang YK. "Dynamic regulation of replication independent deposition of histone H3 in fission yeast." Nucleic Acids Res. 33, 7102-7110, 2005.
- Takayama Y, Takahashi K. "Differential regulation of repeated histones genes during the fission yeast cell cycle." Nucleic Acids Res. 35, 3223-3237, 2007.
- Waterborg JH. "Dynamics of histone acetylation in vivo. A function for acetylation turnover?" Biochem. Cell Biol. 80, 363-378, 2002.
- Cui B, Liu Y, Gorovsky MA. "Deposition and function of histone H3 variants in Tetrahymena thermophila." Mol. Cell. Biol. 26,7719-7730, 2006.
- Smith MM, Santisteban MS, "Genetic dissection of histone function." Methods 15, 269-281, 1998.