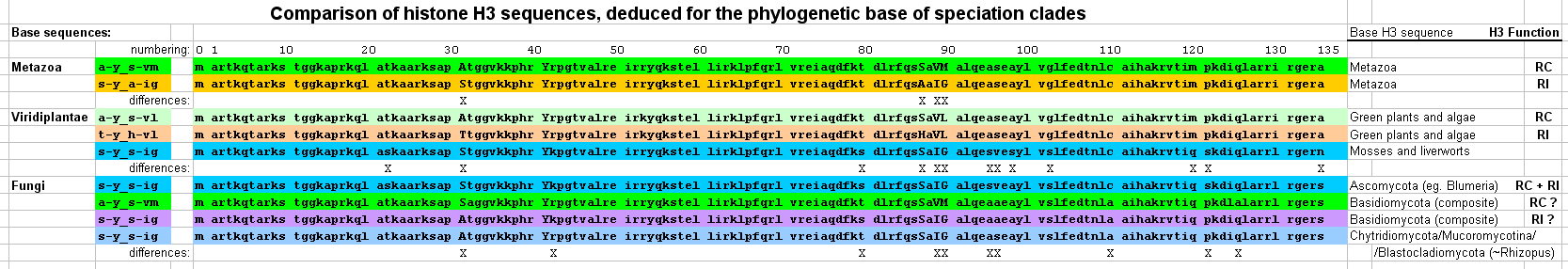
The pattern of histone H3 residues at positions 31, 41, 87, 89 and 90 (named the H3 "idiotype")
is based on sequence differences between histone H3 variants in animals (metazoa) and plants (viridiplantae)
for which distinct Replication-Coupled (RC) nucleosome assembly and
transcription-linked, Replication-Independent (RI) nucleosome assembly pathways have been identified. [1-2]
|
- Metazoan tree (pdf file)
- defines two H3 base sequences with separate H3 variants for RC (a-y_s-vm) and RI (s-y_a-ig) functions.
- Table of all Metazoan H3 sequences compiled.
- Viridiplantae tree (pdf file)
- defines two H3 base sequences with separate H3 variants for RC (a-y_s-vl) and RI (t-y_h-vl) functions, and
- identifies a single H3 histone base (s-y_s-ig) for Mosses and Liverworts.
- Table of all Viridiplantae H3 sequences compiled.
- Fungal tree (pdf file)
- defines a single H3 histone (s-y_s-ig) in Ascomycota with known combined RC and RI functionality,
- defines two H3 base sequences in Basidiomycota which may represent separate H3 variants for RC (a-y_s-vm) and RI (s-y_s-ig) functions, and
- defines a single H3 histone (s-y_s-ig) for a range of major fungal subphyla.
- Table of all Fungal H3 sequences compiled.
|
Phylogenetic Bootstrap Analysis
- Phylogenetic bootstrap analyses using MEGA4 suggest a phylogenetic analysis between the metazoan, fungal and viridiplantae phyla which is inconsistent with the established evolutionary relationship among these groups (see Tree Of Life diagram).
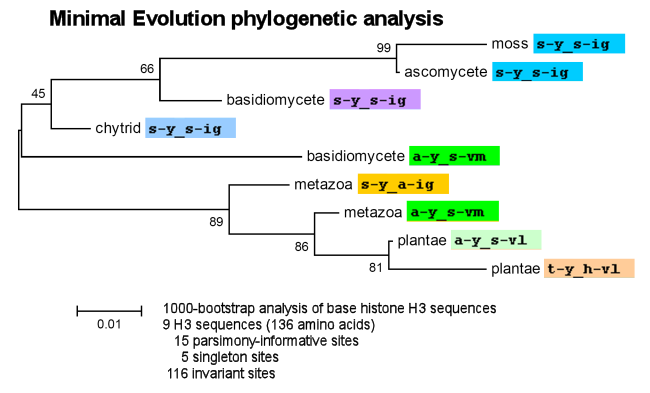
Virtually identical results are obtained in Maximum Parsimony and Neighbor-Joining phylogenetic analyses.
Some variation in branching patterns and bootstrap significance values is obtained with UPGMA.
|
|
|

|
- Histone H3 residues are subject to extremely high purifying selection pressures
that result in the very slow evolution of histone H3 protein forms.
This selection results in two distinct outcomes:
- Amino acid residues required for the general function of histone H3 are invariant to an amazing degree.
Concerted evolution of the histone H3 proteins produced from multiple gene loci is a generally observed fact. [3-4]
If any change occurs in these residues in one of the H3 variants, one may detect co-evolution where the other H3 variant changes in the same way.
Examples:
- In the metazoan tree, both H3 variant forms changed residue 81 from aspartic acid to glutamic acid.
- In the fungal tree, both H3 variants in Filobasidiella changed residue 96 from alanine to serine;
- coordinated changes occurred in Saccharomycetales at 95, in Debaryomyces at 102 and in Candida at 124.
- Amino acid residues required for the RC or RI variant-specific function in nucleosome assembly,
determined to be residues 87, 89 and 90, with the likely addition of residue 31,
remain distinct and invariant over hundreds of millions of years in animals, in plants and in basidiomycetes.
In phyla where the H3 diversity appears to have been reduced to a single histone H3 form,
such as ascomycetes, the sequences are absolutely invariant within the subphylum.
TWO hypotheses could explain RC and RI histone H3 evolution
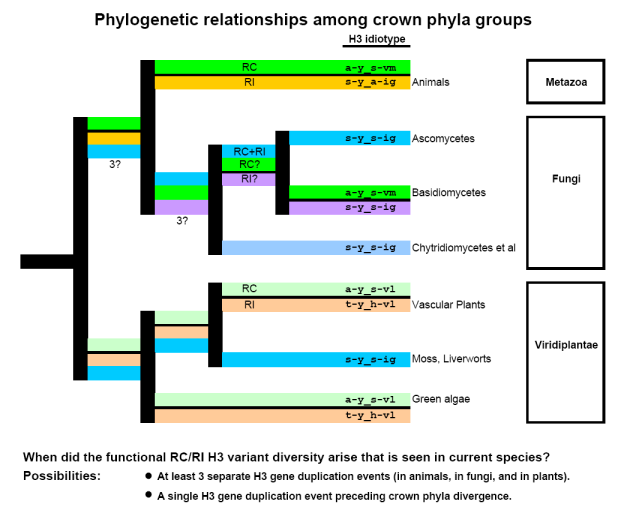
The combination of high selection pressures with a sparsity of parsimony-informative residues across an evolutionary span of 600 million years when animals, fungi and plants became distinct phyla, suggests two possible, mutually exclusive scenarios.
Gene duplication of histone H3 into RC and RI variants:
- H3 gene duplication was an independent event for metazoans, for plants and for basidiomycetes.
This conclusion is supported primarily by formal phylogenetic analyses. [5-6]
- H3 gene duplication preceded the divergence of the crown phyla of animals, plants and fungi.
This conclusion is supported by
- the principle of Maximum Parsimony (a single event explains the data),
- the observation of distinct RC- and RI-nucleosome assembly pathways in ascomycetes with a single H3 form (assumed to result from the loss of the second form after RC- and RI-function support was acquired in a single H3 form), and
- the observation of 2 distinct histone H3 variants with distinct and characteristic H3 idiotypes in protists.
Clades with 2 distinct H3 forms include Chromaveolate (stramenopiles/red algae and archaeplastida) and Alveolata (Tetrahymena, Plasmodium, Cryptosporidium et al). They have not yet been observed in Excavata (Trichomonas, Trypanosoma, et al) or Amoebzoa (Entamoeba, Dictyostelium et al).
The clearest example is Tetrahymena where RC- and RI- functions were identified in differentially expressed H3 variants. [4]
Experimental analysis to evaluate these two possibilities is in progress.
|
|
References
- Ahmad K, Henikoff S. "The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly." Mol. Cell 9, 1191-1200, 2002.
- Henikoff S, Ahmad K. "Assembly of variant histones into chromatin." Annu. Rev. Cell. Dev. Biol. 21, 133-153, 2005.
- Pyne S, Skiena S, Futcher B. "Copy correction and concerted evolution in the conservation of yeast genes." Genetics 170, 1501-1513, 2005.
- Cui B, Liu Y, Gorovsky MA. "Deposition and function of histone H3 variants in Tetrahymena thermophila." Mol. Cell. Biol. 26,7719-7730, 2006.
- Waterborg JH, Robertson AJ. "Common features of analogous replacement histone H3 genes in animals and plants." J. Mol. Evol. 43, 194-206, 1996.
- Thatcher TH, MacGaffey J, Bowen J, Horowitz S, Shapiro DL, Gorovsky MA. "Independent evolutionary origin of histone H3.3-like variants of animals and Tetrahymena." Nucleic Acids Res. 22, 180-186, 1994.
|
|